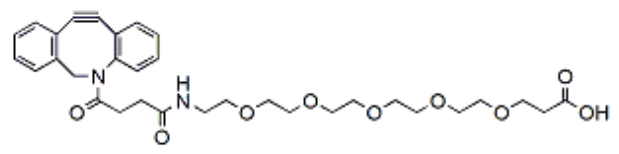
FITC-水杨酸 FITC-salicylic acid 上海金畔生物提供各种CY3,CY5,CY5,CY5.5,CY7,CY7.5荧光标记水杨酸
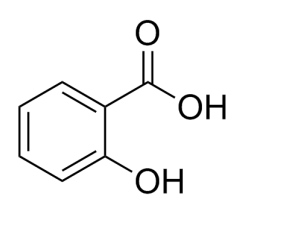
水杨酸-FITC
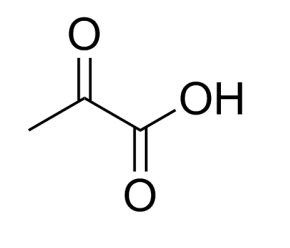
水杨酸名称
中文名 水杨酸
英文名 salicylic acid
中文别名 2-羟基苯甲酸 | 2-羟基苯甲酸 | 柳酸,沙利西酸 | 美沙拉嗪杂质F | 柳酸
英文别名 Verrugon
Acid, Salicylic
Duoplant
Freezone
MFCD00002439
Acid, o-Hydroxybenzoic
水杨酸生物活性
描述 Salicylic acid 抑制 COX-2 活性,抑制作用与转录因子 (NF-κB) 激活无关。
靶点 COX-2 Autophagy Mitophagy
密度 1.4±0.1 g/cm3
沸点 336.3±0.0 °C at 760 mmHg
熔点 158-161 °C(lit.)
分子式 C7H6O3
分子量 138.121
闪点 144.5±19.1 °C
精确质量 138.031693
PSA 57.53000
LogP 2.06
外观性状 白色至灰白色结晶粉末
蒸汽密度 4.8 (vs air)
蒸汽压 0.0±0.7 mmHg at 25°C
折射率 1.616
水溶解性 1.8 g/L (20 ºC)
| 参数信息 | |
|---|---|
| 外观状态: | 固体或粉末 |
| 质量指标: | 95%+ |
| 溶解条件: | 有机溶剂/水 |
| CAS号: | N/A |
| 分子量: | N/A |
| 储存条件: | -20℃避光保存 |
| 储存时间: | 1年 |
| 运输条件: | 室温2周 |
| 生产厂家: | 上海金畔生物科技有限公司 |